Directed cell migration continuum model as explanation for biological pattern formation
During the evolution of multicellularity, cells needed to find a way to self-organise into structures able to fulfil a broad range of physiological functions. Directed cell migration (DCM) is an elegant way to reorder cells and break symmetry based on simple rules and local information, leading to a large variety of density patterns. Hence, developing models capable of describing various forms of directed cell migration is important to achieve a mechanistic understanding of morphogenesis and how organs gain their functional shapes.
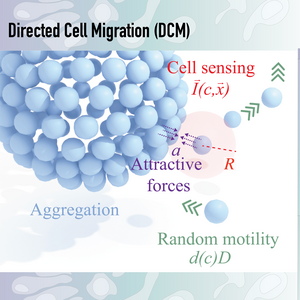
DCM can be described by an partial differential equation system, consisting of a random cell motility therm, similar to Fickian diffusion, and an directed cell motility therm, consisting of an local density kernel, probing the surrounding cell densities and direct cell movement towards high density areas.
Continuum models offer an efficient framework for studying pattern formation at meso- to macro-scales. These models simplify complex cellular behavior and often allow for analytical insight. However, traditional continuum models for DCM have been numerically challenging, especially near domain boundaries, where the local density integrals become difficult to evaluate. Consequently, most implementations have relied on specialized finite volume solvers, limiting their applicability to biologically relevant and irregular geometries.
To address these limitations, we developed a versatile DCM model implementation in COMSOL Multiphysics, leveraging general extrusion operators to handle both periodic and zero-flux boundary conditions in 1D, 2D, and 3D domains. This approach overcomes previous numerical barriers and allows for the simulation of DCM in arbitrary geometries.
Using this framework, we characterized how various model parameters influence the dynamics of pattern formation—such as the speed of pattern emergence, the size of features, and the resulting qualitative shapes. Our robust and flexible COMSOL-based implementation now enables the exploration of a broad range of developmental processes, such as germ layer segregation in gastruloid systems and cartilage ring formation in the developing trachea.
By expanding the toolbox for studying DCM, our work contributes to a deeper understanding of tissue patterning and opens up new possibilities for investigating complex morphogenetic processes in developmental biology.
下载
- Poster_comsol.pdf - 5.25MB
- Mederacke_simulating_comsol_DCM.pdf - 9.58MB
